Reagents
| Abbreviation | Full Name | Specifications |
|---|---|---|
| Tris | Trizma Hydrochloride | Conc. = 1M, pH = 7.5 |
| AA | Acetic Acid | Conc. = 0.5M |
| EDTA | Ethylenediaminetetraacetic Acid | Conc. = 0.1M |
| MA | Magnesium Acetate | Conc. = 0.1M |
| DNA Working Stocks | Various DNA Strands | Conc. = 1 μM |
Procedures for Walker Synthesis
This is the procedure to create the DNA walkers. It is the functional nanoparticle and payload carrier in the system. Depending on the requirements, it can be programmed later to host different attachments such as gold, quantum dots, and so on.
They are prepared from a pre-prepared batch of DNA that has been ordered as programmed strands to assemble the walkers after specific thermocycling. Therefore, the following procedure has to be followed very carefully for the self-assembling phenomenon to occur.
| Time Since Start of Work (hrs.) | Instruction |
|---|---|
| 00:00 | Remove stock solutions of DNA from the Freezer |
| 00:10 | Prepare the buffer solution in a PCR Tube:
|
| 00:30 | Add the following amount of solutions to the PCR Tube (in µL):
|
| 0:40 | Close the PCR tube, tap down any droplets that may be on the sides of the container and vortex for one minute. |
| 0:45 |
|
| 4:45 | Remove the sample from the thermocycler after equilibration, then freeze it. |
Results
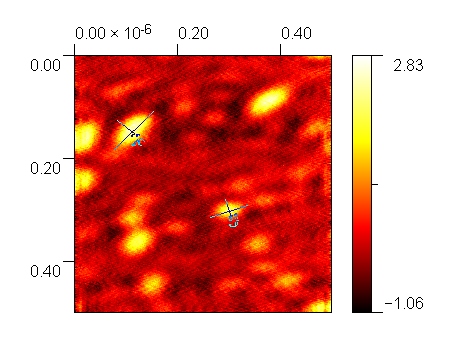
The walkers in this experiment turned out well for the experiment, as they were able to fulfill the purpose of hosting the biotinylated DNA, and were also able to bind to the LGNPs. However, our AFM images show that there was significant aggregation visible in the sample, likely as a result of discrepancies in volumes added, as well as some discrepancies with leaving samples outside the freezer for a long duration. The expected size of the walkers was around 10 nm, but the observed size of the particles according to the AFM images is around 80 - 100 nm.
Reagents
- Subscript denotes solvent type
| Abbreviation | Full Name | Specifications |
|---|---|---|
| Tris | Trizma Hydrochloride | Conc. = 1.0 M, pH = 7.5 |
| AA | Acetic Acid | Conc. = 0.5 M |
| EDTA | Ethylenediaminetetraacetic Acid | Conc. = 100 mM |
| MA | Magnesium Acetate Solution | Conc. = 100 mM |
| DNA | Various DNA Strands | See Table |
| DDIW | Double Deionized Water | R = 14.0 MΩ∙cm-1 |
Procedure
This procedure is for the synthesis of the DNA Origami Cassettes. The Cassettes act as “cargo-loading stations” for the Walkers; the Cassettes donate their nanoparticle cargoes to the Walkers when within a certain proximity.
- The Q Cycler was used to thermo cycle the Cassettes
- The program qCycle.exe was used to run the Q Cycler
- VWR 200 µL PCR tubes were used for the reaction
The following table lists the DNA strands required and the required volumes.
| Strand-ID | Volume (μL) |
|---|---|
| C2-BKL | 10.7 |
| C2-BS | 11 |
| C2-DG | 6.7 |
| C2-SET-J1 | 8.3 |
| C2-SET-J2 | 10.5 |
| C2-PC1 | 9.4 |
| C2-SHIELD | 8.9 |
| C2-LB | 12.4 |
| C2-DR | 8.9 |
| C2 - PC2 | 5.7 |
| C2-P2 | 8 |
| CX-R1 | 9.5 |
| CX-R2 | 9.5 |
| CX-BR | 8.9 |
| CX-P1 | 9.2 |
| CX-BL | 8.3 |
Synthesis Procedure
| Time Since Start of Work (hrs.) | Instruction |
|---|---|
| 00:00 | Take the DNA strands out of the freezer to allow them to thaw |
| 00:05 | Add 8.1 μL of DDIW, 25 μL of MA, 8 μL of Tris, 8 μL AA, 5 μL EDTA |
| 00:10 | Vortex the PCR Tube for 30 seconds, then add all of the required DNA Strands listed in the above table |
| 00:20 | Vortex the PCR Tube for 1 minute, then place in the thermocycler. Close the thermocycler and run the Cassette program, using 150 μL for the volume input |
| 07:20 | Take the PCR tube from the Q Cycler, then store in the freezer after equilibrating to room temperature |
Results
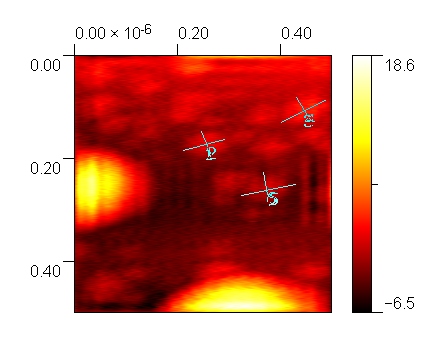
Our AFM characterization results show that the cassettes are approximately 40 nm by 70 nm, which are the desired dimensions. The DNA tile is 150 nm by 200 nm, so 3 cassettes can effectively be fit onto the tile. The cassettes will be conjugated with their respective cargoes before proceeding with walking the walker across the tile.